Bridging Cells and Space: The Journey of scDesign3 Exploration


Dr. Jingyi Jessica Li
Dr. Jingyi Jessica Li, Professor of Statistics and Data Science (also affiliated with Biostatistics, Computational Medicine, and Human Genetics), leads a research group titled the Junction of Statistics and Biology at UCLA. With Ph.D. from UC Berkeley and B.S. from Tsinghua University, Dr. Li focuses on developing interpretable statistical methods for biomedical data. Her research delves into quantifying the central dogma, extracting hidden information from transcriptomics data, and ensuring statistical rigor in data analysis by employing synthetic negative controls. Recipient of multiple awards including the NSF CAREER Ward, Sloan Research Fellowship, ISCB Overton Prize, and COPSS Emerging Leaders Award, her contributions have gained recognition in the fields of computational biology and statistics.
The Article Link:
scDesign3 generates realistic in silico data for multimodal single-cell and spatial omicsRegarding the research background and significance, does this work discover new knowledge or solve existing problems within the field? Please elaborate in detail.
Our work aimed to provide a unified solution to the problems of method benchmarking and parameter inference in the single-cell and spatial omics field. As breakthrough technologies, single-cell and spatial omics can unprecedentedly provide a multimodal view of the molecular biology phenomena (such as genome-wide gene expression and open chromatin regions) in individual cells. The excitement about these new technologies and the generated massive data has propelled a rapid development of computational methods, leading to thousands of computational methods being developed in less than a decade. As a result, the availability of numerous computational methods makes method benchmarking a pressing challenge. Fair benchmarking demands synthetic data that contain ground truths and mimic real data, thus calling for realistic simulators.
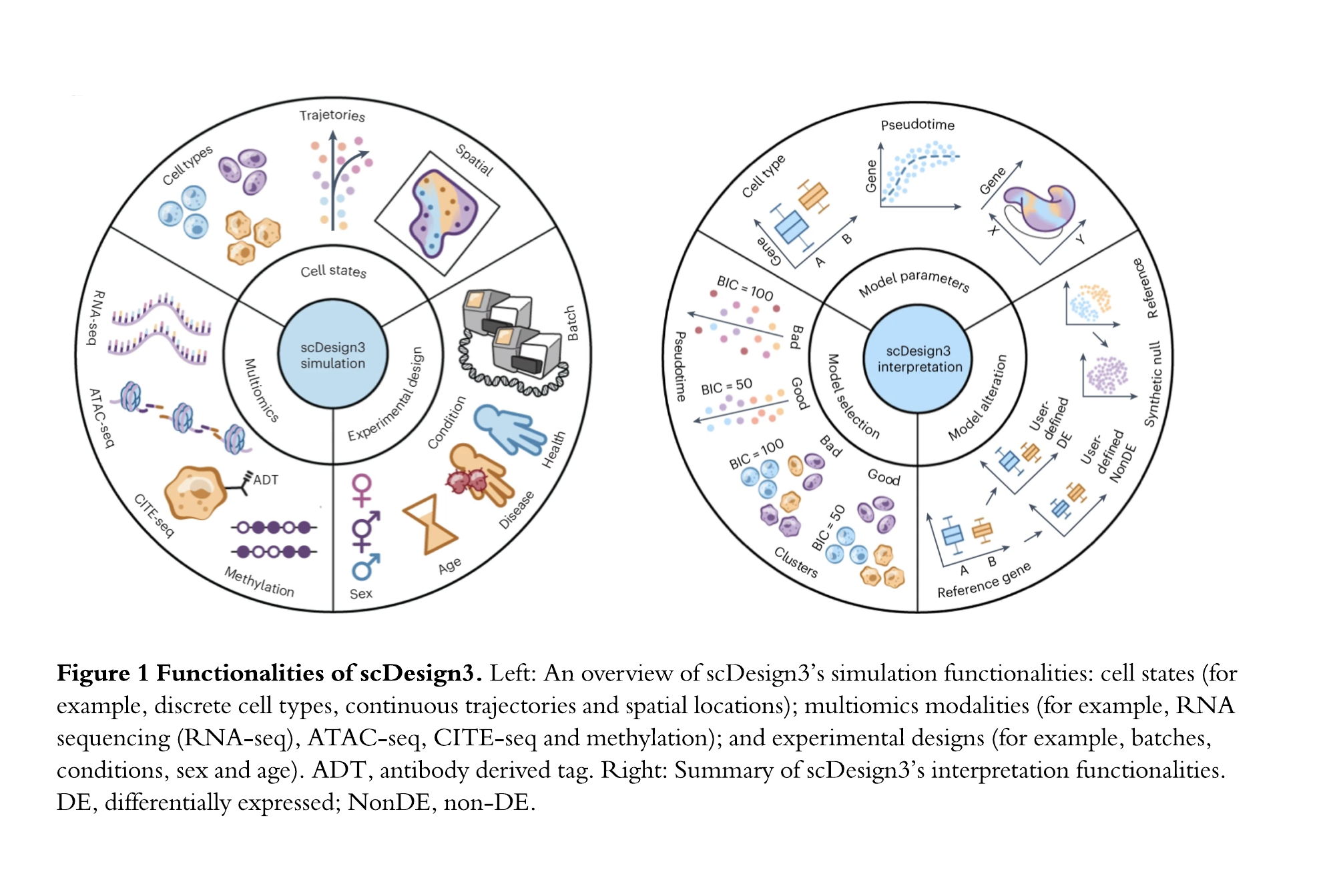
Motivated by the critical need for method benchmarking and other tasks (e.g., power analysis) requiring ground truths, my group has developed a series of mode-based single-cell simulators: scDesign (Li and Li, Bioinformatics 2019) and scDesign2 (Sun et al., Genome Biology 2021), with scDesign3 being the latest version. For each simulator, we propose a statistical model to be fitted to real data to describe a multi-variate distribution of cells; then we simulate synthetic cells by sampling from the fitted distribution. For example, for gene expression data, genes are the variables in the multi-variate distribution; for chromatin accessibility data, genomic regions are the variables. The advance of our simulators was based on the improvement of our proposed model. From scDesign to scDesign2, we added the consideration of variable correlations to make the model fit better to real data; as a result, the synthetic data become more realistic. From scDesign2 to the latest scDesign3, the most comprehensive simulator to our knowledge at the time of publication, we enlarged the model to account for comprehensive scenarios, including various cell latent states (discrete cell types or continuous cell trajectories), omics types, spatial locations, and experimental designs (Figure 1 Left).
An advantage of scDesign3 is that its probabilistic model unifies the generation and inference for single-cell and spatial omics data. The model’s interpretable parameters and likelihood enable scDesign3 to generate customized synthetic data and unsupervisedly assess the goodness-of-fit of cell latent states inferred by a computational method (Figure 1 Right). Hence, scDesign3 is not only a simulator that can generate realistic synthetic data but also a statistical model that can help interpret real experimental data. By inferring the parameters of the scDesign3 model, we can perform hypothesis testing tasks such as identifying the genes whose expression changes between cell types or along a cell trajectory, as well as comparing gene correlations between cell types.
How did the reviewers evaluate (praise) it?
The reviewers acknowledged the timeliness and significance of our work. Their quotes include “Overall, this simulator is timely and invaluable for benchmarking computational methods and interpreting single-cell and spatial omics data.” “The current manuscript scDesign3 proposed a substantial extension from the author’s previous work and addressed some of the key concerns with many current simulation studies; in particular, the capturing of the correlation structure between genes. This manuscript has two key components. The first is the focus on the creation of a generalizable simulation framework and the second component illustrates how the scDesign3 can be applied in three different modelling settings, (i) parameter estimation and inference, (ii) model selection, and (iii) perturbation).”
If this achievement has potential applications, what are some specific applications it might have in a few years?
I think realistic synthetic data generation holds promise for critical applications in the single-cell and spatial omics field. First, I have been advocating the inclusion of synthetic negative control data to help users discern spurious discoveries made from a complex analysis pipeline. Such discoveries often arise from artifacts generated by the pipeline due to the “double-dipping” issue, which leads to the confirmatory bias. Second, synthetic data can potentially help alleviate the data imbalance issue, which can negatively affect tasks such as data integration and supervised learning. Third, synthetic data with added perturbations can be used for evaluating the stability of computational results. Given the increasing complexity of data analysis pipelines, involving many user-specified heuristic thresholds across multiple steps, it is essential to ensure the robustness of computational results.
Can you recount the specific steps or stages from setting the research topic to the successful completion of the research?
I will answer this question based on the way we generally conduct research. Before setting a research topic, we always think about three aspects: significance and novelty, validation approaches, and target user base. Given the competitive nature and widespread interest in single-cell and spatial omics, a comprehensive literature review is the cornerstone to justify the novelty and significance of the research topic. Equally important is the deliberation on how to validate the efficacy of our proposed method, whether through computational analysis or experimental verification. We must assess the availability of resources for validation and identify potential collaborators if such resources are lacking. Moreover, we must consider the prospective end-users of our method: will our method be useful for biologists who generate single-cell and spatial omics data, or will it only be interesting to method developers like ourselves? Importantly, how can we persuade biologists, who typically rely on visualization and empirical data, of the utility of our method? In general, I think answering these questions is essential to advocate a computational method to the biomedical scientists.
Were there any memorable events during the research? You can tell a story about anything related to people, events, or objects.
An interesting story is about the term “real data,” used by statisticians in almost every methodology paper. As the scDesign projects were all about “synthetic data,” a term juxtaposed with “real data,” I initially used the term “real data” when introducing our scDesign work to biologists. Surprisingly, some biologists did not understand the concept of "real data," asking questions such as "What defines real data?" and "Is there such a thing as fake data?" This confusion stemmed from the presumption that all data were generated from experiments and hence qualify as real. Aligned with this confusion, many biologists cannot appreciate the utility of simulation, considering simulation as purely a decoration of a computational method because simulation always works. I think realizing and recognizing these differences in mindsets (e.g., abstract thinking vs. empirical thinking) is an essential first step in fostering collaborations with biomedical scientists.
Is there a follow-up plan based on this research? If so, please elaborate.
We plan to develop an interactive software package for real-data-based synthetic data generation and applications. We will make the package compatible with the state-of-the-art single-cell analysis pipelines Seurat in R and Scanpy in Python. By providing users with the capability to generate customized synthetic data, we hope our software will enable users to benchmark computational methods on their specific datasets, leading to more rigorous and trustworthy discoveries.
Without a doubt, AI is one of the hot topics of 2023, requiring extensive data support in its development. What assistance can biostatistics offer to the development of AI?
I view AI as a technology advance, similar to the advent of computers in the previous century. Statistics transitioned from its pre-computer era, where mathematical methods were the sole tools, to the computer era, where statistical analyses leveraged computing power to tackle previously impossible missions. Regardless of the tool—from mathematics to computers—the core of statistics, how to infer hidden truths from observed data, remains a timeless pursuit. Therefore, I am optimistic that the ongoing development of AI will pave the way for unprecedented advancements in statistical methodologies. I envision a synergy between the feature extraction capabilities of AI and the inferential toolkits of statistics, which can together propel data-driven scientific discovery.
Besides the above questions, is there anything else about this achievement that you would like to add? If so, please add it below.
I believe statisticians can play a pivotal role in elucidating the impact of proper versus improper use of statistics on scientific discoveries. Demonstrating the impact on concrete examples could significantly enhance the perceived value and influence of statistics within the scientific community.
Proofread by: Hongtu Zhu